Dr. Steven Mielke

Steve Mielke is a research scientist in the Austin, Texas area. He is a former programmer and analyst at NASA, NASA Postdoctoral Program fellow, NIH National Research Service Award recipient, and AAAS Mass Media fellow.
Professional Certifications and Specializations
• Machine Learning
• IBM Data Science
• Deep Learning
View My GitHub Profile
spmielke@gmail.com linkedin.com/in/steven-mielke/
Protein Structure Estimation from Data Mining
Project Description
Summary
Motivated by the need for fast, reliable methods of assessing protein structure in the era of big genomics, this continuing effort seeks to provide data-derived tools for high-throughput structural characterization of proteins directly from traditional (e.g., HSQC) nuclear magnetic resonance (NMR) spectra, prior to resonance assignment. By correlating the NMR chemical shifts and structure content of proteins of known conformation, these tools allow accurate, real-time estimation of secondary structure in previously uncharacterized proteins.
Key projects contributing to this effort have 1) empirically correlated the averaged chemical shift (ACS) of protein backbone nuclei with observed secondary structure (α‑helix and β‑strand) content as a fraction of overall conformation;1-3 2) confirmed these results by applying rigorous statistical methods to further correlate ACS with protein structural classes;3-5 and 3) extended the latter work by introducing a neural network model that improves the accuracy and specificity of structural class estimation from ACS values.6
Current projects further explore deep learning strategies, and apply other machine learning approaches, such as logistic regression analysis, to new and expanded datasets.7
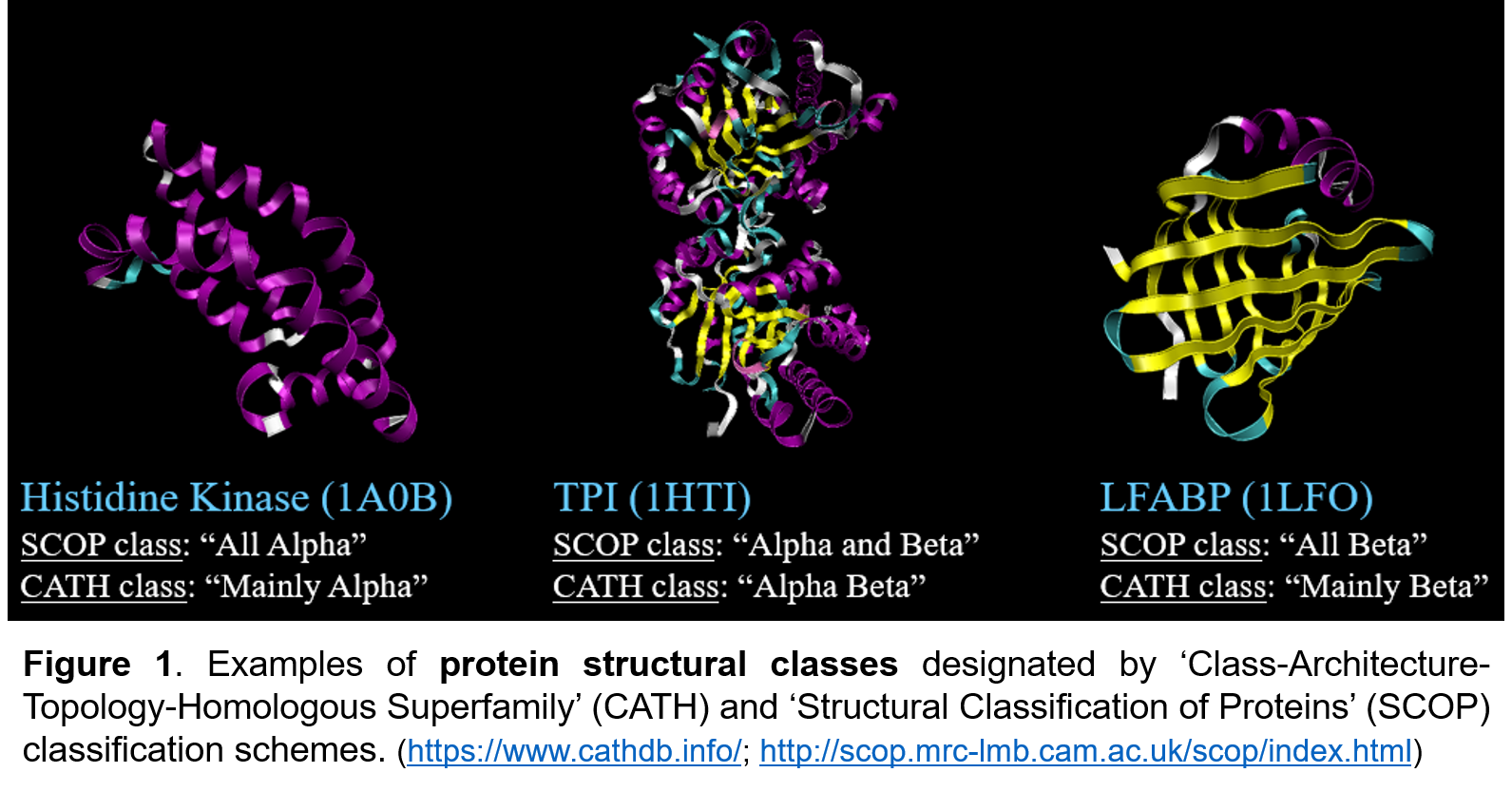
(See Repository for selected project files.)
References
-
Sibley, A.B., Cosman, M. and Krishnan, V.V. (2003) An empirical correlation between secondary structure content and averaged chemical shifts in proteins. Biophys. J. 84, 1223–1227.
-
Mielke, S.P. and Krishnan, V.V. (2005) Estimation of protein secondary structure content directly from NMR spectra using an improved empirical correlation with averaged chemical shift. Journal of Structural and Functional Genomics 6, 281–285.
-
Mielke, S.P. and Krishnan, V.V. (2008) Characterization of protein secondary structure from NMR chemical shifts. Progress in NMR Spectroscopy 54, 141–165.
-
Mielke, S.P. and Krishnan, V.V. (2003) Chemical shift-based identification of protein structural classes. Biophysical Journal 84, 460A.
-
Kumar, A.V., Rehana, F.M.A., Cao, Y. and Krishnan, V.V. (2015) Application of data mining tools for classification of protein structural class from residue based averaged NMR chemical shifts. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 1854, 1545-1552.
-
Mielke, S.P. In preparation.